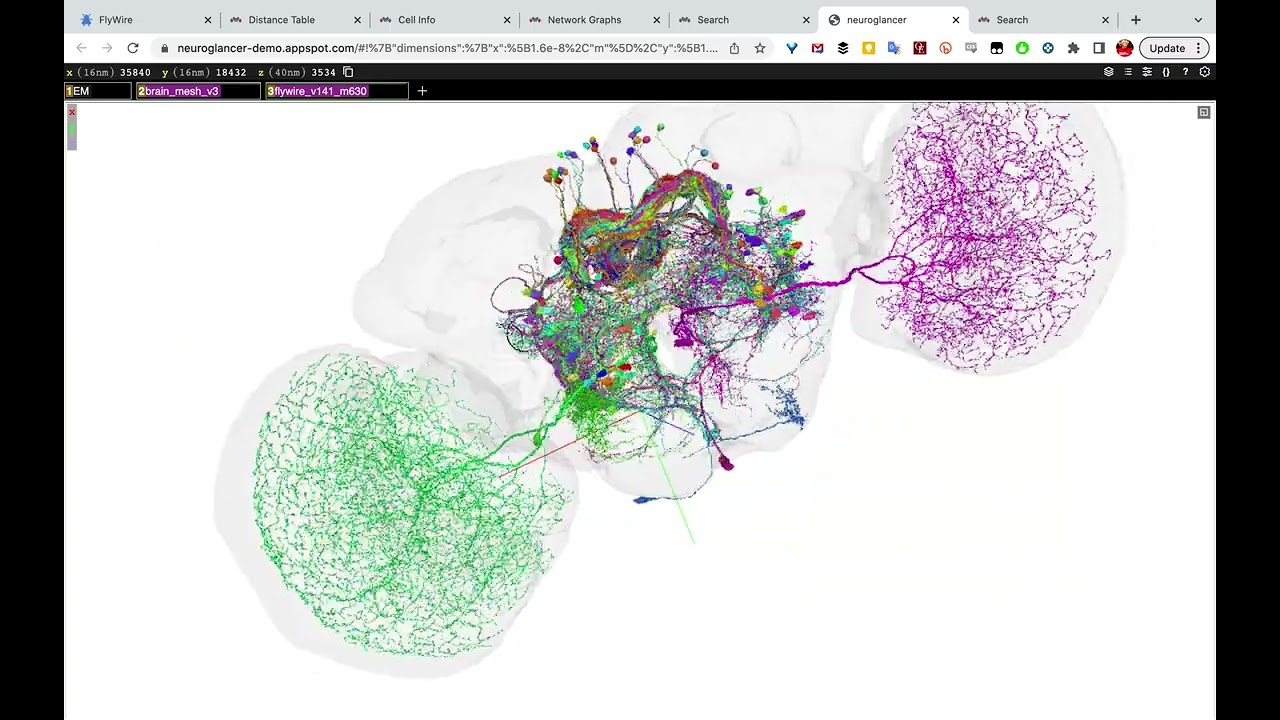
On 2023-06-30 the FlyWire Consortium released a whole-brain connectome of the female fruit fly Drosophila melanogaster. This is the “wiring diagram” of the fly’s brain, including 130,000 neurons, tens of millions of synapses, and 75 million connections, annotated and typed.
The work is presented in two preprints published on bioRχiv, listed below with their abstracts.
“Neuronal wiring diagram of an adult brain”
Connections between neurons can be mapped by acquiring and analyzing electron microscopic (EM) brain images. In recent years, this approach has been applied to chunks of brains to reconstruct local connectivity maps that are highly informative, yet inadequate for understanding brain function more globally. Here, we present the first neuronal wiring diagram of a whole adult brain, containing 5\times 10^7 chemical synapses between ~130,000 neurons reconstructed from a female Drosophila melanogaster. The resource also incorporates annotations of cell classes and types, nerves, hemilineages, and predictions of neurotransmitter identities. Data products are available by download, programmatic access, and interactive browsing and made interoperable with other fly data resources. We show how to derive a projectome, a map of projections between regions, from the connectome. We demonstrate the tracing of synaptic pathways and the analysis of information flow from inputs (sensory and ascending neurons) to outputs (motor, endocrine, and descending neurons), across both hemispheres, and between the central brain and the optic lobes. Tracing from a subset of photoreceptors all the way to descending motor pathways illustrates how structure can uncover putative circuit mechanisms underlying sensorimotor behaviors. The technologies and open ecosystem of the FlyWire Consortium set the stage for future large-scale connectome projects in other species.
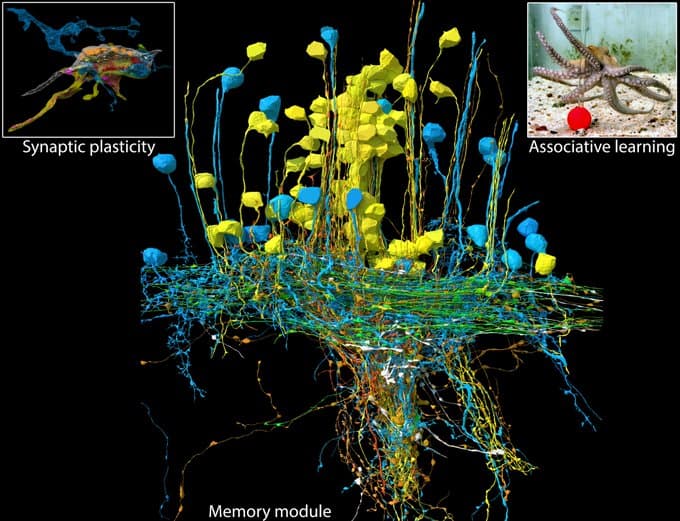
The fruit fly Drosophila melanogaster combines surprisingly sophisticated behaviour with a highly tractable nervous system. A large part of the fly’s success as a model organism in modern neuroscience stems from the concentration of collaboratively generated molecular genetic and digital resources. As presented in our FlyWire companion paper, this now includes the first full brain connectome of an adult animal. Here we report the systematic and hierarchical annotation of this ~130,000-neuron connectome including neuronal classes, cell types and developmental units (hemilineages). This enables any researcher to navigate this huge dataset and find systems and neurons of interest, linked to the literature through the Virtual Fly Brain database. Crucially, this resource includes 4,179 cell types of which 3,166 consensus cell types are robustly defined by comparison with a second dataset, the “hemibrain” connectome. Comparative analysis showed that cell type counts and strong connections were largely stable, but connection weights were surprisingly variable within and across animals. Further analysis defined simple heuristics for connectome interpretation: connections stronger than 10 unitary synapses or providing >1% of the input to a target cell are highly conserved. Some cell types showed increased variability across connectomes: the most common cell type in the mushroom body, required for learning and memory, is almost twice as numerous in FlyWire than in the hemibrain. We find evidence for functional homeostasis through adjustments of the absolute amount of excitatory input while maintaining the excitation-inhibition ratio. Finally, and surprisingly, about one third of the cell types recorded in the hemibrain connectome could not be robustly identified in the FlyWire connectome, cautioning against defining cell types based on single connectomes. We propose that a cell type should be robust to inter-individual variation, and therefore defined as a group of cells that are more similar to cells in a different brain than to any other cell in the same brain. We show that this new definition can be consistently applied to whole connectome datasets. Our work defines a consensus cell type atlas for the fly brain and provides both an intellectual framework and open source toolchain for brain-scale comparative connectomics.
Access to this massive database is via “Codex: Connectome Data Explorer”, which is freely available to anybody (Google login required), and provides a variety of ways to explore and analyse the data, as well as the ability to download raw data for offline analyses. A video gallery of examples of using the Codex is available. The following is an overview of Codex.