“Synthia”, or Mycoplasma laboratorium, is a synthetic bacterium created by the J. Craig Venter Institute by starting with the parasitic bacterium Mycoplasma mycoides and stripping down its genome to find the minimal set of genes that would allow metabolism and reproduction. (This is not unlike Madman Muntz removing removing parts from TV sets to find the cheapest minimal circuit that would work—for a while—a process dubbed “Muntzing”. The search for a minimal genome is reminiscent of Spiegelman’s Monster, although that did not produce a free-living organism and required an external supply of RNA polymerase to reproduce.) After arriving at a genome of 493 genes with 541,560 base pairs, they synthesised DNA encoding the genome, inserted it in a bacterium from which its own DNA had been removed, and observed their new organism, dubbed JCVI-syn3A, reproduce and metabolise. This work is described in the 2019 eLife paper, “Essential metabolism for a minimal cell”. Here is the abstract.
JCVI-syn3A, a robust minimal cell with a 543 kbp genome and 493 genes, provides a versatile platform to study the basics of life. Using the vast amount of experimental information available on its precursor, Mycoplasma mycoides capri, we assembled a near-complete metabolic network with 98% of enzymatic reactions supported by annotation or experiment. The model agrees well with genome-scale in vivo transposon mutagenesis experiments, showing a Matthews correlation coefficient of 0.59. The genes in the reconstruction have a high in vivo essentiality or quasi-essentiality of 92% (68% essential), compared to 79% in silico essentiality. This coherent model of the minimal metabolism in JCVI-syn3A at the same time also points toward specific open questions regarding the minimal genome of JCVI-syn3A, which still contains many genes of generic or completely unclear function. In particular, the model, its comparison to in vivo essentiality and proteomics data yield specific hypotheses on gene functions and metabolic capabilities; and provide suggestions for several further gene removals. In this way, the model and its accompanying data guide future investigations of the minimal cell. Finally, the identification of 30 essential genes with unclear function will motivate the search for new biological mechanisms beyond metabolism.
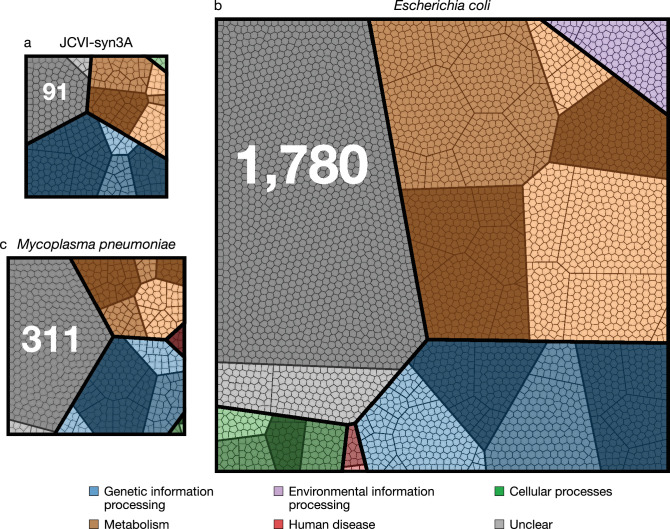
Here is a comparison of the genome of JCVI-syn3A with two other well-known bacteria, with genes colour-coded by function.
The original JCVI-syn3A was a static organism, incapable of movement. Now, by adding two genes extracted from Spiroplasma eriocheiris, a crustacean pathogen, researchers have created an organism, syn3Bsw, which is capable of swimming, albeit in a different manner and not as far as the source bacterium. This work is described in the November 2022 paper in Science Advances, “Reconstitution of a minimal motility system based on Spiroplasma swimming by two bacterial actins in a synthetic minimal bacterium”, with abstract:
Motility is one of the most important features of life, but its evolutionary origin remains unknown. In this study, we focused on Spiroplasma, commensal, or parasitic bacteria. They swim by switching the helicity of a ribbon-like cytoskeleton that comprises six proteins, each of which evolved from a nucleosidase and bacterial actin called MreB. We expressed these proteins in a synthetic, nonmotile minimal bacterium, JCVI-syn3B, whose reduced genome was computer-designed and chemically synthesized. The synthetic bacterium exhibited swimming motility with features characteristic of Spiroplasma swimming. Moreover, combinations of Spiroplasma MreB4-MreB5 and MreB1-MreB5 produced a helical cell shape and swimming. These results suggest that the swimming originated from the differentiation and coupling of bacterial actins, and we obtained a minimal system for motility of the synthetic bacterium.
